Watching SARS-CoV-2 Spread in Animal Models in Real Time Using New “Reporter Viruses”
0 View
Share this Video
- Publish Date:
- 7 October, 2021
- Category:
- Covid
- Video License
- Standard License
- Imported From:
- Youtube
Tags
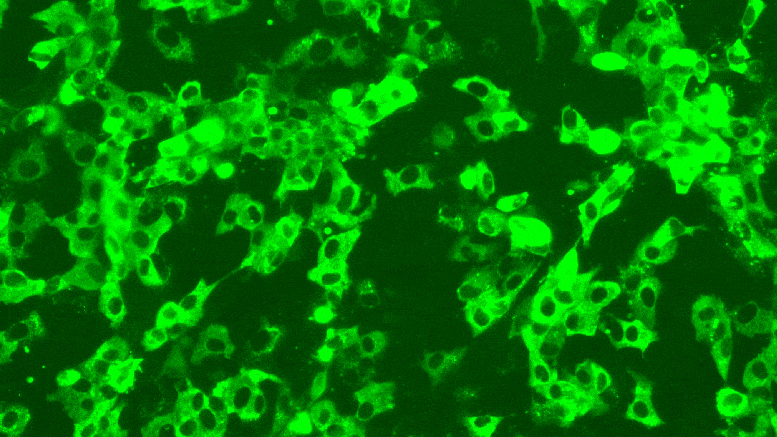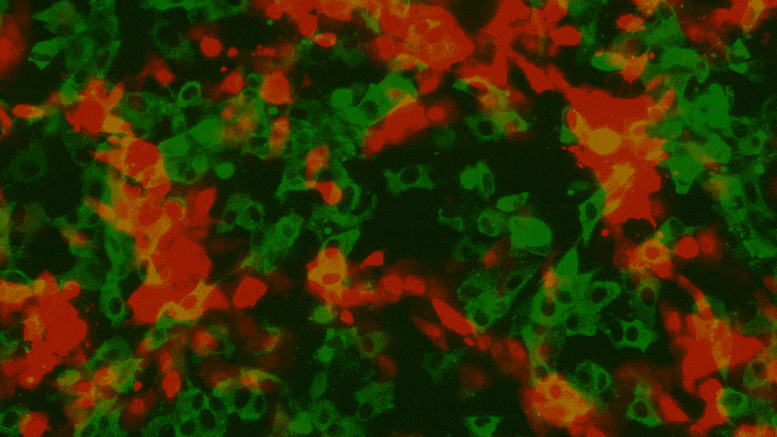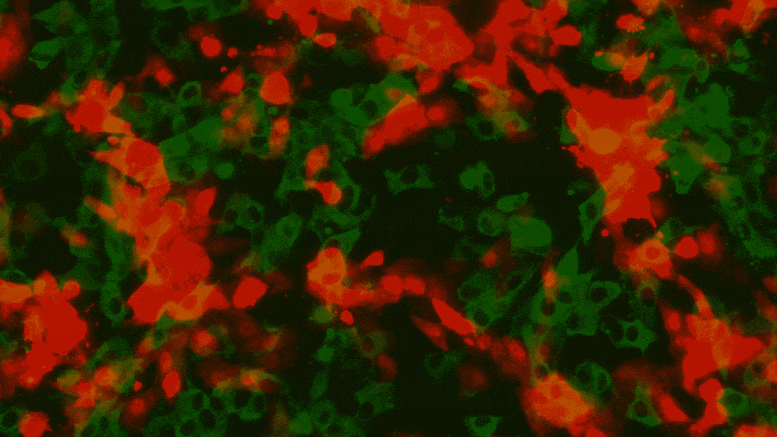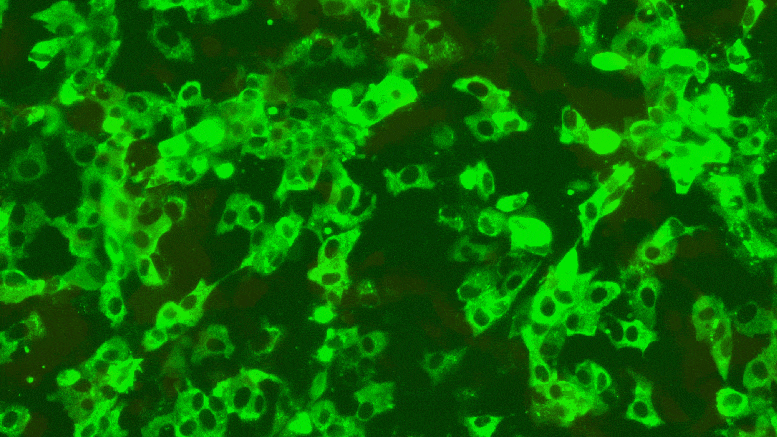
Texas Biomed researchers have developed reporter viruses to express different colors for different variants of SARS-CoV-2. This way they can easily see whether treatments, vaccines or neutralizing antibodies against multiple variants work at the same time. Credit: Texas Biomed
New “reporter viruses” developed by Texas Biomed researchers make it much easier to detect SARS-CoV-2 and its variants in cells and live animals in the lab, and enable faster screening of potential antiviral drugs, vaccines and neutralizing antibodies .
A version of SARS-CoV-2, the virus that causes the disease COVID-19, has been successfully modified to glow brightly in cells and animal tissues, providing a real-time way to track the spread and intensity of viral infection like that. occurs in animal models, researchers at the Texas Biomedical Research Institute (Texas Biomed) report in the journal The Proceedings of the National Academy of Sciences (PNAS).
“Now we can track where the virus is going in animal models for COVID-19,” said virologist Luis Martinez-Sobrido, Ph.D., a Texas Biomed professor and senior paper author. “Being able to see how the virus is progressing and which organs and cell types it specifically targets will be of great help in understanding the virus and optimizing antiviral drugs and vaccines.”
Texas Biomed Researchers Kevin Chiem, Ph.D. candidate (left), and Chengjin Ye, Ph.D. (right) Prepare to analyze non-infectious samples of SARS-CoV-2 in a fluorescent imaging machine in the lab of Texas Biomed Professor Luis Martinez-Sobrido, Ph.D. Credit: © Billy Calzada/San Antonio Express-News via ZUMA Press Wire
In addition to monitoring the virus, Martinez-Sobrido and his collaborators have already started using the reporter viruses to screen for how well neutralizing antibodies work against different variants of concern, as reported recently in the Journal of Virology.
Turn on the lights
To create the reporter virus, Martinez-Sobrido and his team combined several advanced molecular biology tools to add the genetic sequence for the fluorescent or bioluminescent “reporter” proteins to the virus’s genetic code. As the virus’s code is replicated and transcribed, so is the code for the glowing proteins.
In a previous study, the team replaced one of the virus’s genes with the gene for the glowing proteins, but this resulted in a very weak signal — the gene was not expressed enough to be easily detected in animals. To increase the brightness, the researchers had to figure out how the virus could produce higher amounts of the reporter proteins.
Left: In a previous study, Texas Biomed researchers attempted to exchange a viral gene with a green fluorescent protein gene, but that did not result in enough proteins being expressed. Right: In this study, they placed the fluorescent protein gene next to the most highly expressed protein of SARS-CoV-2, and it worked very well. Each green spot reports one viral particle. Credit: Texas Biomed
Their solution: they placed the reporter gene next to another gene in SARS-CoV-2, namely the gene encoding the nucleocapsid protein. “It’s the protein most expressed in SARS-CoV-2,” says molecular biologist Chengjin Ye, Ph.D., a member of Martinez-Sobrido’s lab. This time the signal was so bright, “it almost blinded me when I looked through the fluorescent microscope,” he said.
Faster screens
The reporter proteins work in cells and living animal models, in conjunction with imaging systems that detect the wavelengths of light emitted by the proteins. Being able to visually observe viral load and location offers many advantages over other methods. It is much simpler and faster, which saves time and material.
The new reporter viruses help researchers observe the progression of SARS-CoV-2 infection in transgenic mouse models. Numbers indicate days after infection and red indicates a greater amount of virus replication. Here, the amount of viral load reported in the lungs increases on day 2 after infection and then decreases again on day 6 after infection. Credit: Texas Biomed
“Instead of needing a large team to screen 2,000 compounds to see if they work against the virus, one person could do that in a few hours with a reporter virus,” Ye said.
It also makes it possible to monitor the virus in the same animal during infection and treatment, reducing the number of animals needed to get comparable insights.
Follow variants
The team modified the reporter viruses to express different colored proteins attached to SARS-CoV-2 variants of concern, which they described in a separate article in the Journal of Virology. Crucially, this approach has allowed them to test how well a neutralizing antibody works against two variants in one test well at the same time.
Professor Luis Martinez-Sobrido, Ph.D. Credit: Texas Biomed
“This is a significant advantage in saving time and resources, especially with so many feedstocks such as plastics and reagents that are in such high demand and limited supply due to the pandemic,” said Kevin Chiem, Ph.D. candidate and member of Martinez-Sobrido’s lab. “If new variants emerge, we can easily adapt the system and quickly screen for how well antibodies work against them.”
Powerful and accurate
Importantly, the group showed that the reporter viruses behave similarly to a wild-type version of the virus. This is due to the fact that they didn’t delete any viral genes and because they designed the reporter protein to immediately separate from the virus’s nucleocapsid protein so that it functions normally. Their study shows that reporter protein brightness correlates well with viral load, although protein accumulation can occur over several days, leading to a slightly stronger signal in some cases.
The advances are based on several powerful techniques, including reverse genetics techniques to generate recombinant SARS-CoV-2, which link pieces of genetic code together to produce the entire virus.
Martinez-Sobrido and his team have shared their recombinant SARS-CoV-2 and its non-infectious precursor materials, called plasmids, with more than 100 labs around the world. They can now share the reporter viruses with qualified labs with biocontainment safety level (BSL)-3 access, which is needed to work with SARS-CoV-2 to help fight the ongoing COVID-19 pandemic.
“We feel it is our responsibility to share these new tools and technologies with other researchers around the world to end the pandemic as quickly as possible,” said Martinez-Sobrido.
Reference: “Analysis of SARS-CoV-2 Infection Dynamics in Vivo Using Reporter Expressing Viruses” by Chengjin Ye, Kevin Chiem, Jun-Gyu Park, Jesus A. Silvas, Desarey Morales Vasquez, Julien Sourimant, Michelle J Lin, Alexander L. Greninger, Richard K. Plemper, Jordi B. Torrelles, James J. Kobie, Mark R. Walter, Juan Carlos de la Torre, and Luis Martinez-Sobrido, September 24, 2021, Proceedings of the National Academy of Sciences .
DOI: 10.1073/pnas.2111593118
Contributors to these projects include Jun-Gyu Park, Jesus A. Silvas, Desarey Morales Vasquez, and Jordi B. Torrelles at Texas Biomed; Julien Sourimant and Richard K. Plemper at The Center for Translational Antiviral Research at Georgia State University; Michelle J. Lin and Alexander L. Greninger at the University of Washington; James J. Kobie, Mark R. Walter and Michael S. Piepenbrink at the University of Alabama at Birmingham; and Juan Carlos de la Torre of the Scripps Research Institute.










