Supercomputer Simulations Reveal How Dominant COVID-19 Strain Binds to Host, Succumbs to Antibodies
0 View
Share this Video
- Publish Date:
- 13 May, 2021
- Category:
- Covid
- Video License
- Standard License
- Imported From:
- Youtube
Tags
Supercomputer simulations at Los Alamos National Laboratory have shown that the G form of SARS-CoV-2, the dominant strain of the virus that causes COVID-19, has mutated into a conformation that allows it to attach more easily to host receptors while also is more sensitive to antibodies than the original D form. Credit: Los Alamos National Laboratory
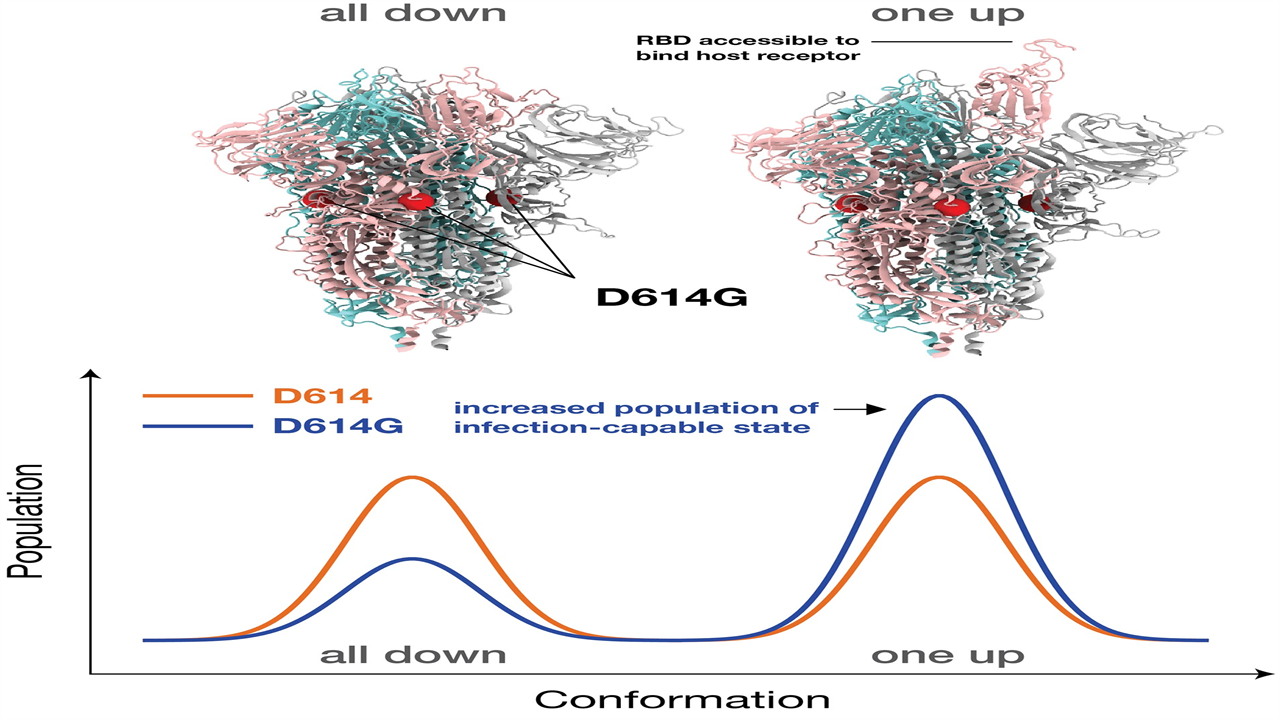
The dominant G-shaped Spike protein is more likely to ‘stick’ its head up to adhere to receptors, but that makes it more vulnerable to neutralization.
Large-scale atomic-level supercomputer simulations show that the dominant G-shape variant of the COVID-19-causing virus is more contagious, in part due to its greater ability to easily bind to the target host receptor in the body, compared to other variants. These research results from a team led by Los Alamos National Laboratory highlight the mechanism of both G-form infection and antibody resistance to it, which could aid in the future development of vaccines.
“We found that the interactions between the basic building blocks of the Spike protein become more symmetrical in the G-shape, giving it more opportunities to bind to the receptors in the host – in us,” said Gnana Gnanakaran, corresponding author. of the paper recently published in Science Advances. “But that also means that antibodies can neutralize it more easily. Essentially, the variant pops up to bind to the receptor, giving antibodies a chance to attack it. “
Researchers knew the variant, also known as D614G, was more contagious and could be neutralized by antibodies, but they didn’t know how. By simulating more than a million individual atoms and requiring approximately 24 million CPU hours of supercomputer time, the new work provides molecular-level details about the Spike’s behavior of this variant.
Current vaccines for SARS-CoV-2, the virus that causes COVID-19, are based on the original D614 form of the virus. This new understanding of the G variant – the most comprehensive supercomputer simulations of the G shape at the atomic level – could mean it provides a backbone for future vaccines.
The team discovered the D614G variant in early 2020, as the COVID-19 pandemic caused by the SARS-CoV-2 virus was growing. These findings are published in Cell. Scientists had observed a mutation in the Spike protein. (In all variants, it is the Spike protein that gives the virus its characteristic corona.) This D614G mutation, named after the amino acid at position 614 in the SARS-CoV-2 genome that underwent a substitution for aspartic acid, was prevalent worldwide. a matter of weeks.
The Spike proteins bind to a specific receptor found in many of our cells through the Spike’s receptor binding domain, ultimately leading to infection. That binding requires the receptor binding domain to change structurally from a closed conformation, which cannot bind, to an open conformation, which can.
The simulations in this new research show that interactions between the building blocks of the Spike are more symmetrical in the new G-shaped variant than those in the original D-shaped trunk. That symmetry leads to more viral spikes in the open conformation so that it can infect a person more easily.
A team of postdoctoral fellows from Los Alamos – Rachael A. Mansbach (now assistant professor of physics at Concordia University), Srirupa Chakraborty, and Kien Nguyen – led the research by conducting multiple microsecond-scale simulations of the two variants in both conformations. of the receptor binding domain to illustrate how the Spike protein interacts with both the host receptor and the neutralizing antibodies that can help protect the host against infection. The research team also included Bette Korber of Los Alamos National Laboratory and David C. Montefiori of Duke Human Vaccine Institute.
The team would like to thank Paul Weber, Head of Institutional Computing at Los Alamos, for providing access to the lab’s supercomputers for this study.
Reference: “The SARS-CoV-2 Spike variant D614G prefers an open conformational state” by Rachael A. Mansbach, Srirupa Chakraborty, Kien Nguyen, David C. Montefiori, Bette Korber, S. Gnanakaran, April 16, 2021, Science Advances.
DOI: 10.1126 / sciadv.abf3671
Funding: The project was supported by Los Alamos Laboratory Directed Research and Development project 20200706ER, Director’s Postdoctoral fellowship, and the Center of Nonlinear Studies Postdoctoral Program in Los Alamos.










