Scientists Go “Inside” the COVID-19 Viral Protein To Attack a Weak Point
0 View
Share this Video
- Publish Date:
- 17 November, 2021
- Category:
- Covid
- Video License
- Standard License
- Imported From:
- Youtube
Tags
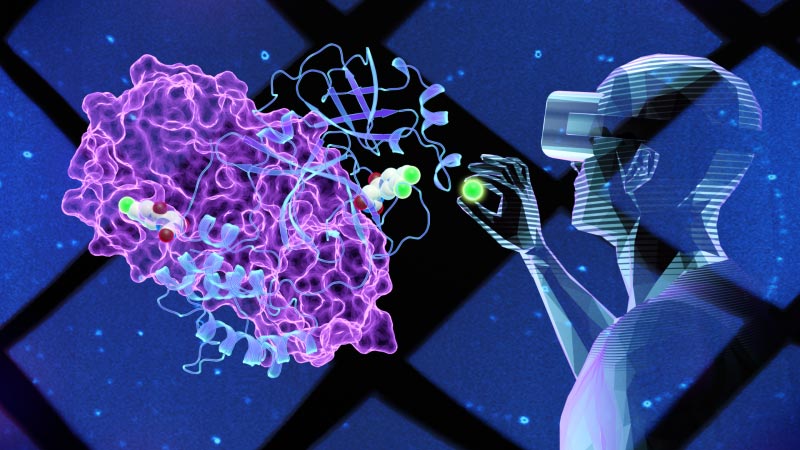
Virtual reality technology has allowed scientists to look ‘inside’ the Covid-19 virus and develop a new molecule that can inhibit the main protease enzyme. Credit: Jill Hemman/ORNL
Virtual reality (VR) technology allows scientists to create 3D models of an object and then virtually “go inside” to look around to better understand its structure and function.
This is what researchers at the Department of Energy (DOE) Oak Ridge National Laboratory (ORNL) did to study the SARS-CoV-2 virus that caused the COVID-19 pandemic. The team used neutrons and X-rays to map some of the internal structure of the coronavirus to create an accurate 3D model. In particular, the scientists mapped the major protease (Mpro), an enzyme involved in virus replication, to which they added a preliminary small molecule discovered using rapid computer screening.
Using VR to look at the enzyme model, the scientists virtually constructed several small molecules by modifying their structures to see if newly designed compounds could fit or bind to a key site on the Mpro enzyme surface. A strong enough binding can inhibit or block the enzyme’s functioning, which is vital to prevent the virus from multiplying in patients with COVID-19.
To determine the effects of specific chemical modifications on the binding of the 19 inhibitor candidates to the Mpro enzyme, the team synthesized each inhibitor molecule and measured their binding strengths. The stronger the bond, the more effectively the inhibitor would block the action of the enzyme and the replication of the virus.
One of the test inhibitors, labeled HL-3-68, showed a superior ability to bind to and inhibit the function of Mpro compared to others tested. Details of the study, titled “Structural, electronic and electrostatic determinants for inhibitor binding to subsites S1 and S2 in SARS-CoV-2 main protease,” are published in the Journal of Medicinal Chemistry.
“Our study aimed to better understand how molecules bind to the active site of the Mpro enzyme, which plays a key role in SARS-CoV-2 replication,” said lead author Daniel Kneller. “While testing the molecules we designed, we discovered one with a single extra chlorine atom that showed a greater ability to inhibit Mpro. This new chemical structure is different from what has been previously studied by the global community and could open up new avenues of research.” can open up exciting possibilities for combating SARS-CoV-2.”
The active site on the Mpro enzyme is common in other types of coronaviruses and does not appear to mutate easily, providing an opportunity to potentially design an antiviral treatment that works against multiple SARS-CoV-2 variants and other coronaviruses.
Equally importantly, the active site is different from those known in human enzymes, which would minimize the potential for inadvertent binding that could lead to side effects in patients.
The X-ray measurements and production of the Mpro enzyme samples were performed by the Center for Structural and Molecular Biology using facilities at ORNL’s Spallation Neutron Source (SNS) and resources at the High Flux Isotope Reactor (HFIR). The inhibitor candidates were synthesized by co-authors Hui Li and Peter Bonnesen of the Macromolecular Nanomaterials group of the Center for Nanophase Materials Sciences (CNMS).
“This study combined a plethora of biophysical, biochemical and molecular biology methods, and involved virtual reality-assisted structural analysis and small-molecule building, involving scientists from across ORNL, Argonne National Laboratory, the National Institutes of Health and the University of Tennessee were brought together.-Knoxville.The collaborative nature of the study allowed us to uncover the rules that small molecule inhibitors must obey when they bind to the enzyme to be useful for further steps in the long process of drug design and development,” said corresponding author Andrey Kovalevsky.
Co-corresponding author Peter Bonnesen added: “This was a new and exciting project for the CNMS to work on, and it drew on our expertise in synthesizing custom organic molecules for our users. For this project, we provided our SNS colleagues from a few candidate molecules at a time. When the results came back regarding the molecules’ effectiveness as inhibitors, the team would discuss what adjustments to make to the molecular structure. Then Hui and I would go back to the lab to make these to create new candidate inhibitors.”
The study also shed light on the ability of the Mpro enzyme to change its shape and change its electrical charge from positive to negative, or from negative to positive, depending on the size and structure of the inhibitor molecule it binds to. These features are important to understand when developing an effective inhibitor molecule.
To investigate neutron scattering, the scientists used the macromolecular neutron diffractometer (MaNDi) at the SNS for its ability to collect data from the relatively small samples the team had to work with.
“Due to the urgent nature of the research related to the SARS-CoV-2 virus, we were only able to grow relatively small samples of the Mpro enzyme,” said co-author Leighton Coates. “Because smaller samples scatter neutrons weakly and this results in ‘noisy’ neutron data, data analysis can be difficult. The time structure of the neutron beam at the MaNDi instrument allowed us to remove most of the noise, increasing the signal-to-noise ratio, thus we have a lot more actionable data to work with.”
Next steps for the ORNL researchers include testing chemical modifications of the HL-3-68 inhibitor to determine whether newly designed compounds can bind even better than HL-3-68 to more effectively inhibit and ultimately kill the Mpro enzyme. prevent the coronavirus from replicating.
Meanwhile, the researchers made their data public through the Protein Data Bank to more quickly inform and assist the world’s scientific and medical communities. Of course, more research and testing is needed to validate the effectiveness and safety of an inhibitor as a COVID-19 treatment. However, this study could provide an opportunity for other scientists to conduct additional research that would benefit billions of people around the world.
Reference: “Structural, electronic, and electrostatic determinants for inhibitor binding to subsites S1 and S2 in SARS-CoV-2 master protease” by Daniel W. Kneller, Hui Li, Stephanie Galanie, Gwyndalyn Phillips, Audrey Labbé, Kevin L. Weiss, Qiu Zhang, Mark A. Arnould, Austin Clyde, Heng Ma, Arvind Ramanathan, Colleen B. Jonsson, Martha S. Head, Leighton Coates, John M. Louis, Peter V. Bonnesen and Andrey Kovalevsky, Oct. 27, 2021, Journal of Medicinal Chemistry .
DOI: 10.1021/acs.jmedchem.1c01475
Other co-authors of the paper include Stephanie Galanie, Gwyndalyn Phillips, Audrey Labbé, Kevin L. Weiss, Qiu Zhang, Mark A. Arnould, Austin Clyde, Heng Ma, Arvind Ramanathan, Colleen B. Jonsson, Martha S. Head and John M. Louis. Hugh O’Neill of ORNL assisted with sample preparation.
The COVID-19 research at ORNL was supported in part by the Office of Science’s National Virtual Biotechnology Laboratory, a consortium of national DOE labs focused on responding to COVID-19, with funding from the Coronavirus CARES Act. This work was also supported by the National Institutes of Health’s National Institute of Diabetes and Digestive and Kidney Diseases.










