Scientists Catch Shape-Shifting Coronavirus Protein Complex in the Act
0 View
Share this Video
- Publish Date:
- 4 August, 2021
- Category:
- Covid
- Video License
- Standard License
- Imported From:
- Youtube
Tags
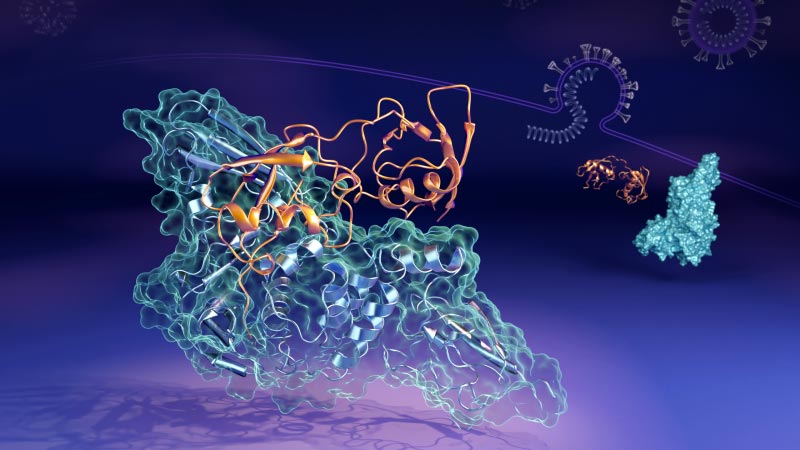
ORNL researchers found that the papain-like protease (in orange) can bind to the human interferon-stimulated gene 15 protein (in blue) in multiple ways and shapes. Credit: ORNL/Jill Hemman
While all viruses have a means of fighting the body’s immune system, scientists have been investigating how the SARS-CoV-2 coronavirus — the cause of the global COVID-19 pandemic — can bypass the immune system in humans.
Now, scientists at the U.S. Department of Energy’s (DOEs) Oak Ridge National Laboratory (ORNL) have revealed the molecular details of how a key protein (the virus’s papain-like protease, or “PLpro”) binds to form a paired structure, or “complex”, with a human protein called interferon-stimulated gene 15 (ISG15). PLpro strips ISG15 from other human cellular proteins to help SARS-CoV-2 evade the immune response. Understanding how the two proteins interact can help develop therapeutic drug treatments that prevent their formation and allow a person’s immune system to better fight off the invading virus.
The study results, titled “Conformional Dynamics in the Interaction of SARS-CoV-2 Papain Like Protease with Human Interferon-Stimulated Gene 15 Protein,” were published in the Journal of Physical Chemistry Letters.
“In human cells infected by the virus, the PLpro of the SARS-CoV-2 virus tends to seek out and bind to the ISG15 protein, an important part of the cells’ immune response,” says Hugh O’Neill, leader of ORNL’s Bio-Facilities group and director of the lab’s Center for Structural Molecular Biology. “When PLpro binds to ISG15, it causes the ISG15 to change shape. The main finding is that the ISG15 can take multiple forms when it binds to PLpro.”
Using small-angle neutron scattering (SANS) at ORNL’s High Flux Isotope Reactor (HFIR), the researchers were able to study the changes in the complex as they occurred.
“We improved the contrast between the PLpro and ISG15 by creating PLpro in which many of the hydrogen atoms have been replaced by deuterium atoms,” said Kevin Weiss, an expert in biodeuteration. “Neutrons interact differently with deuterium atoms, so this helped us better distinguish between the two proteins.
“We used neutrons to analyze the complex in solution, which better simulates the actual physiological environment of the human body,” said Leighton Coates, instrument systems science and technology manager for ORNL’s Second Target Station. “This allowed us to study the changing shapes of the complex that other techniques could not have picked up.”
“The information we gained from our experiments expands our understanding of how the virus works and allows us to build more accurate computer models for other scientists to use,” said Wellington Leite, lead author and postdoctoral researcher at ORNL. “Researchers can use the model to quickly search for sites on the ISG15 that the PLpro attaches to and then try to block those sites.”
Susan Tsutakawa, a biochemist staff scientist at Lawrence Berkeley National Laboratory (Berkeley Lab), obtained small-angle X-ray scattering (SAXS) data on the PLpro-ISG15 complex in the Berkeley Lab’s Advanced Light Source Synchrotron. “In the SAXS studies, we were able to separate different complexes in the sample by coupling SAXS to size-exclusion chromatography while obtaining higher resolution data of the overall configuration of the complex, complementing the SANS studies examining the revealed conformations of individual components in the complex,” Tsutakawa said.
The team plans to conduct additional experiments on this type of biological complex to investigate how small molecules can block the binding of PLpro to ISG15.
Reference: “Conformational dynamics in the interaction of SARS-CoV-2 papain-like protease with human interferon-stimulated gene 15 protein” by Wellington C. Leite, Kevin L. Weiss, Gwyndalyn Phillips, Qiu Zhang, Shuo Qian, Susan E Tsutakawa, Leighton Coates, and Hugh O’Neill, June 10, 2021, Journal of Physical Chemistry Letters.
DOI: 10.1021/acs.jpclett.1c00831
This research was supported by the DOE National Virtual Biotechnology Laboratory, a consortium of national DOE labs funded by the Coronavirus CARES Act. Additional support was provided by the DOE Office of Science, Berkeley Lab’s Molecular Biophysics and Integrated Bioimaging Division, and ORNL’s Center for Structural Molecular Biology.
HFIR is a DOE Office of Science user facility. UT-Battelle LLC manages ORNL for the DOE Office of Science. The Office of Science is the largest proponent of basic science research in the United States and is working to address some of the most pressing challenges of our time.










