Potential New Treatment Target Discovered in the Fight Against COVID-19
0 View
Share this Video
- Publish Date:
- 24 May, 2021
- Category:
- Covid
- Video License
- Standard License
- Imported From:
- Youtube
Tags
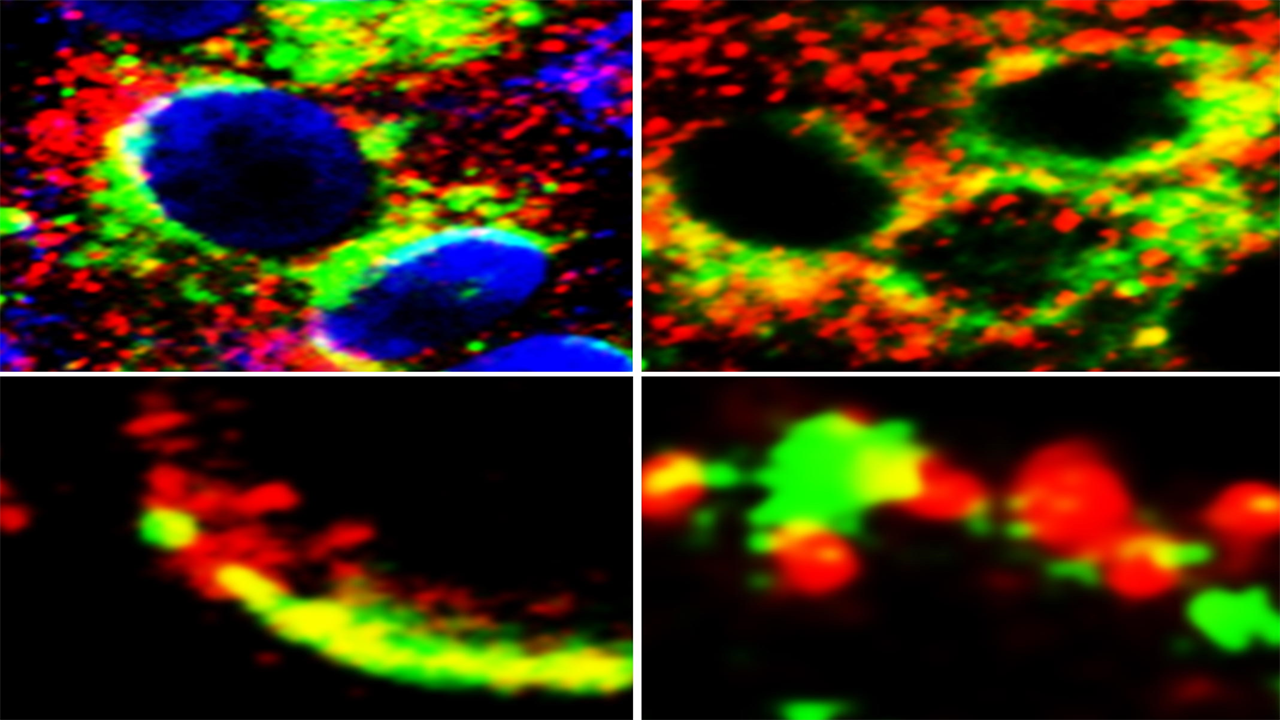
Images of cells showing proximity (yellow) of GRP78 (green) with SARS-Cov-2 Spike protein (red) (left panels) and ACE2 (red) (right panels). Credit: Lead author Anthony Carlos, PhD
USC researchers’ Keck School of Medicine identify a new target and a potential new therapy that could provide an additional treatment option against COVID-19 and some of its variants.
The rapid development of vaccines has provided an essential tool to combat the spread of the deadly SARS-CoV-2 virus, but the challenge of achieving herd immunity is the emergence of new mutations and the inability of immunosuppressed humans In order to develop an effective immune response after vaccination indicates the need for additional solutions to maximize protection.
A new USC study published in the Journal of Biological Chemistry reveals how therapies targeting a molecular chaperone called GRP78 may provide additional protection against COVID-19 and other coronaviruses emerging in the future.
Chaperones such as GRP78 are molecules that help regulate the correct folding of proteins, especially when a cell is under stress. But in some cases, viruses can hijack these chaperones to infect target cells, where they reproduce and spread. GRP78 is involved in the spread of other serious viruses, such as Ebola and Zika.
GRP78 plays more than one role in COVID-19
While studies have shown that SARS-CoV-2, the virus that causes COVID-19, infects cells by binding to ACE2 receptors on their surface, researchers at the Keck School of Medicine of USC investigated whether GRP78 also plays a role.
They found that GRP78 serves as a co-receptor and stabilizer between ACE2 and SARS-CoV-2, improving recognition of the virus’s spike protein and enabling more efficient viral access to host cells.
Senior authors Amy S. Lee, PhD (left), Keigo Machida, PhD (center), Parkash Gill, MD (right), Keck School of Medicine of USC. Credit: Photos courtesy of Don Milici and Richard Carrasco
This study provides the first experimental evidence to support computer modeling predictions, showing that GRP78 binds the SARS-CoV-2 Spike protein in cells. Interestingly, computer modeling further shows that COVID-19 variants that are more infectious bind more strongly to GRP78.
In addition, the research team found that GRP78 also binds to and acts as a regulator of ACE2 – bringing the protein to the cell surface, providing SARS-CoV-2 with more points to bind with and infect cells.
“Our study shows that therapy targeting GRP78 could be more effective in protecting and treating people who have contracted COVID-19 than vaccines alone, especially when it comes to people who cannot receive the vaccine and variants that can bypass vaccine protection. yet depend on GRP78 for access and production, ”said senior author Amy S. Lee, PhD, Judy and Larry Freeman Chair of Basic Science and Professor in the Department of Biochemistry and Molecular Medicine at USC’s Keck School of Medicine.
How SARS-CoV-2 hijacks GRP78
GRP78’s job as a chaperone molecule is to fold proteins in the endoplasmic reticulum (ER), a protein production plant. In stress, including stress caused by SARS-CoV-2 infection, GRP78 is sent to the cell surface. There, it facilitates the binding between ACE2 and Spike protein of SARS-CoV-2, leading to enhanced viral access. Once inside the cell, viruses are known to hijack the ER protein folding machine, of which GRP78 is a key player, to produce more viral proteins.
This process can be intensified in cells under stress from other diseases such as diabetes or cancer, which may be one of the reasons why people with underlying health conditions are more susceptible to SARS-CoV-2 infection.
To investigate the role of GRP78 in SARS-CoV-2 infection, researchers treated lung epithelial cells with humanized monoclonal antibody (hMAb159), which is known to remove GRP78 from the cell surface without adverse effects in mouse models. Intervention removed GRP78 and reduced cell surface ACE2, reducing the number of targets to which SARS-CoV-2 could attach.
These findings led researchers to conclude that interventions, such as hMAb159, to remove cell surface GRP78 can reduce SARS-CoV-2 infection and inhibit the spread and severity of COVID-19 in infected humans.
Potential for targeted treatment on GRP78
Healthy cells require a fraction of GRP78 to function normally. However, stressed cells, such as virally infected or cancer cells, need more GRP78 to survive and multiply, so treatments that reduce the amount of GRP78 in the body can reduce the severity of SARS-CoV-2 infection and spread without adverse effects .
Although this study used a monoclonal antibody, researchers say there are other agents that can be used to reduce the amount or activity of GRP78, creating multiple pathways for potential drug solutions to target GRP78.
“What’s especially exciting about this finding is that GRP78 could be a universal target in combination with existing therapies to combat not only COVID-19 but other deadly viruses that rely on GRP78 for infectivity,” said Lee .
The next step for the research team is to further investigate these findings through animal studies.
Reference: “The GRP78 chaperone is a host helper for SARS-CoV-2 and GRP78-depleting antibodies blocking viral access and infection” by Anthony J. Carlos, Dat P. Ha, Da-Wei Yeh, Richard Van Krieken, Chun-Chih Tseng , Pu Zhang, Parkash Gill, Keigo Machida, and Amy S. Lee, May 2021, Journal of Biological Chemistry.
DOI: 10.1016 / j.jbc.2021.100759
About the study
This study was a collaborative effort of expert researchers in the basic sciences, virology and experimental therapies. Researchers took advantage of USC’s Biosafety-Level 3 containment laboratory, which allowed them to safely study the role of GRP78 on SARS-CoV-2 infection using live virus cultures.
In addition to Lee, the study authors include Parkash Gill, MD, of the department of medicine; Keigo Machida, PhD, and Da-Wei Yeh, PhD, from the Department of Molecular Microbiology and Immunology; and lead author Anthony Carlos, PhD, Dat Ha, Richard Van Krieken, PhD, Chun-Chih Tseng, PhD, and Pu Zhang from the Department of Biochemistry and Molecular Medicine.
This study was supported by grants from the National Institute of Health (R01 CA027607, R01 CA238029, R01 CA027607-37S1, R01 AA025204-01A1, R21 AA025470-01A1 and P50 AA11999) and a pilot grant from a gift from the WM Keck Foundation to support COVID-19 research.










