New Findings on COVID-19 Evolution Using Novel Technology Could Inform Treatment and Vaccine-Development Efforts
0 View
Share this Video
- Publish Date:
- 18 April, 2021
- Category:
- Covid
- Video License
- Standard License
- Imported From:
- Youtube
Tags
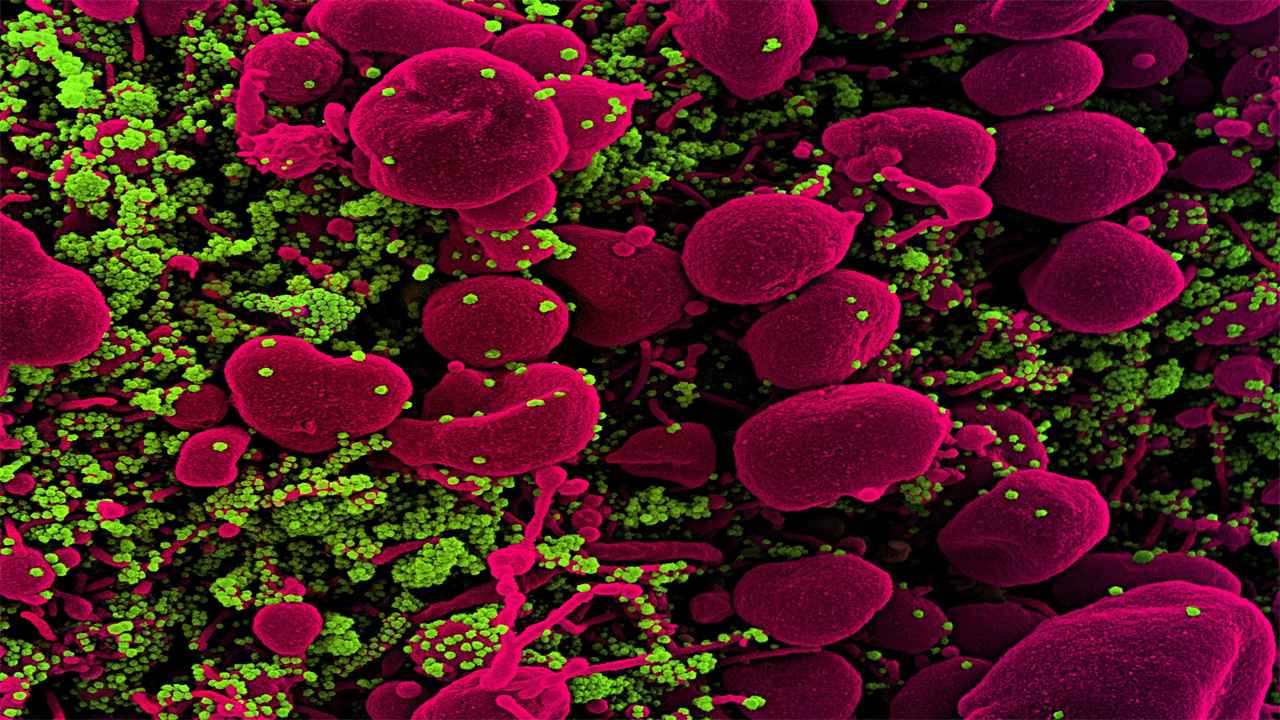
Stained scanning electron micrograph of an apoptotic cell (pink) heavily infected with SARS-COV-2 virus particles (green) isolated from a patient sample. Image taken at the NIAID Integrated Research Facility (IRF) in Fort Detrick, Maryland. Credit: National Institute of Allergy and Infectious Diseases / NIH
Two studies support the key role for the immune system in shaping SARS-CoV-2 evolution
Two studies published in the open access journal PLOS Pathogens provide new evidence supporting an important role for the immune system in shaping the evolution of SARS-CoV-2, the virus that causes COVID-19. These findings – and the new technology behind them – improve understanding of how new SARS-CoV-2 strains emerge, which could help guide treatment and vaccination efforts.
For the first study, Rachel Eguia of Fred Hutchinson Cancer Research Center in Seattle, Washington, and colleagues sought to better understand SARS-CoV-2 by examining a closely related virus that has been widely distributed for a much longer time: cold virus 229E.
229E and SARS-CoV-2 both belong to the coronavirus family, which contains a “spike protein” that allows infection of human cells. A person infected with 229E develops an immune response to the spike protein that protects them from reinfection, but only for a few years. Whether reinfection then occurs because the immune response decreases or because 229E evolves to escape it was unclear.
Eguia and colleagues established this question by testing the activity of serum samples collected from patients in the 80s-90s against spike proteins from both old 229E strains and strains that evolved later. They found that the old spike proteins were vulnerable to the older sera. However, modern spike proteins were able to evade older sera, while remaining vulnerable to sera from modern patients.
This analysis suggests that modern strains of 229E have collected spike protein mutations that allow them to evade older sera. These findings raise the possibility that SARS-CoV-2 and other coronaviruses could undergo a similar evolution, and that COVID-19 vaccines may need to be periodically updated to remain effective against new strains.
The authors add, “The human common cold coronavirus evolves over years to decades to erode neutralization by human polyclonal serum antibodies. This work suggests that human coronaviruses undergo significant antigenic evolution that may contribute to eventual reinfection. “
For the second study, Sung Hee Ko of the National Institute of Allergy and Infectious Diseases in Bethesda, Maryland, and colleagues developed new technology for genetic sequencing of the SARS-CoV-2 spike protein, allowing detection of multiple SARS-CoV-2 strains that may be present simultaneously in a single infected patient.
Previous studies have used standard sequencing methods to produce a single genetic sequence from an individual patient, obscuring the possible presence of multiple SARS-CoV-2 strains. In contrast, the new technology emphasizes virus diversity within each patient and makes it possible to monitor the evolution of new SARS-CoV-2 strains during acute infection.
Indeed, when the researchers applied the new method to human respiratory samples, they found new SARS-CoV-2 variants that arose in the same patient during an acute infection. The precise mutations in these variants suggest that they arose in response to selective pressure from the immune system.
Future application of the new technology could improve understanding of how the evolution of new SARS-CoV-2 variants within a single patient affects their outcomes. The findings also suggest that patients may see greater benefits from early treatment with antivirals that can target multiple strains than from delayed treatment with a single antiviral.
The authors add, “We have used new technology to demonstrate that coronavirus variants with mutated spike proteins can arise early in the course of infection. Our results suggest more virus evolution in each person than previously thought, with potential implications for clinical outcomes and the emergence of transmissible variant strains. “
Together, these two studies deepen the understanding of how new SARS-CoV-2 strains arise in response to immune system activity, potentially paving the way for additional research and improved treatment.
References:
“High-throughput, single-copy sequencing reveals SARS-CoV-2 spike variants coinciding with increasing humoral immunity during acute COVID-19” by Sung Hee Ko, Elham Bayat Mokhtari, Prakriti Mudvari, Sydney Stein, Christopher D. Stringham, Danielle Wagner, Sabrina Ramelli, Marcos J. Ramos-Benitez, Jeffrey R. Strich, Richard T. Davey Jr., Tongqing Zhou, John Misasi, Peter D. Kwong, Daniel S. Chertow, Nancy J. Sullivan, and Eli A. Boritz , April 8, 2021, PLOS Pathogens.
DOI: 10.1371 / journal.ppat.1009431 “A human coronavirus evolves antigen to escape antibody immunity” by Rachel T. Eguia, Katharine HD Crawford, Terry Stevens-Ayers, Laurel Kelnhofer-Millevolte, Alexander L. Greninger, Janet A. Englund, Michael J. Boeckh and Jesse D. Bloom, April 8, 2021, PLOS Pathogens.
DOI: 10.1371 / journal.ppat.1009453
Funding (Paper 1): Funding was provided by the Intramural Research Program of the US National Institutes of Health (project AI005157-01, EAB). MJR-B. is supported by NIGMS Postdoctoral Research Associate Training Program (1FI2GM137804-01). The funders had no role in the design of the research, the collection and analysis of data, the decision to publish or the preparation of the manuscript.
Funding (Paper 2): This work was supported by the following grants from the National Institute of Allergy and Infectious Disease of the National Institutes of Health (https://www.niaid.nih.gov/): R01AI127893 (to JDB), R01AI141707 (to JDB), F30AI149928 (to KDC). JDB is an investigator at Howard Hughes Medical Institute (HHMI, https://www.hhmi.org/). The funders had no role in the design of the research, the collection and analysis of data, the decision to publish or the preparation of the manuscript.










