New Device Can Diagnose COVID-19 and Variants From Saliva Samples
0 View
Share this Video
- Publish Date:
- 6 August, 2021
- Category:
- Covid
- Video License
- Standard License
- Imported From:
- Youtube
Tags
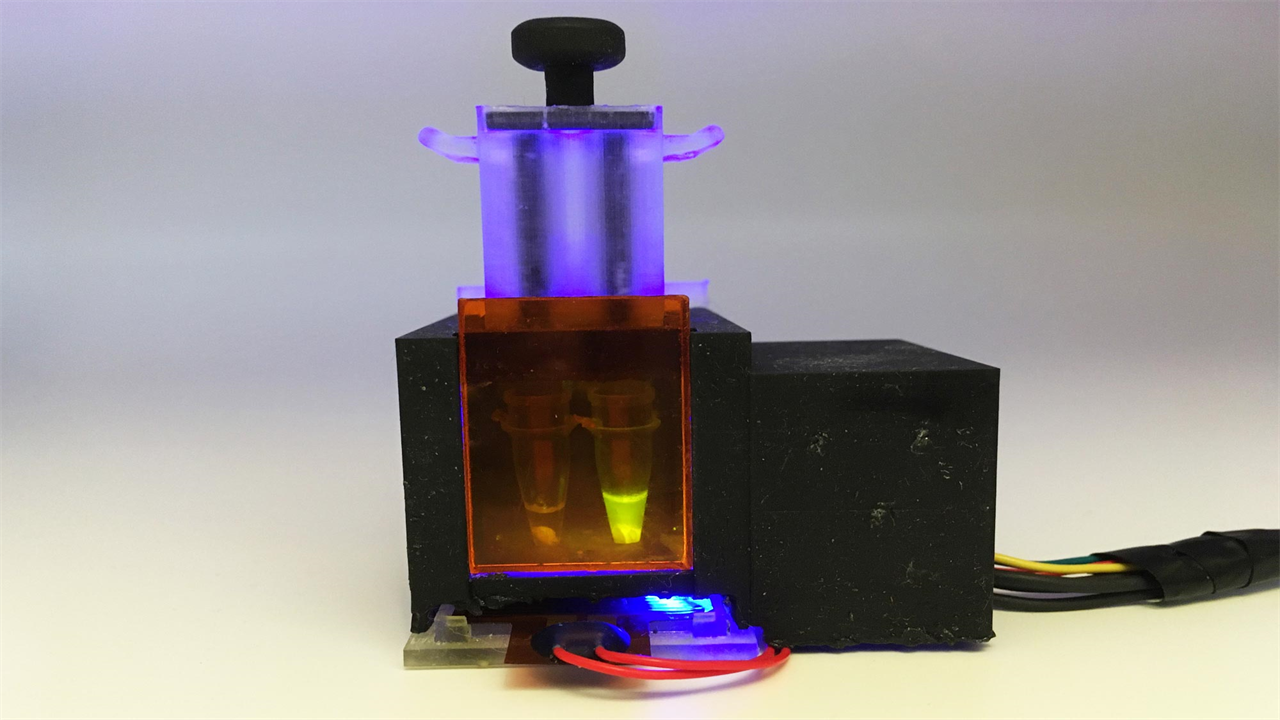
Engineers at MIT and Harvard University have designed a small tabletop device that can detect SARS-CoV-2 from a saliva sample in about an hour. Credit: Devora Najjar
The tabletop diagnosis provides results within an hour and can be programmed to detect variants of the SARS-CoV-2 virus.
Engineers at MIT and Harvard University have designed a small tabletop device that can detect SARS-CoV-2 from a saliva sample in about an hour. In a new study, they showed that the diagnosis is just as accurate as the PCR tests now used.
The device can also be used to detect specific viral mutations associated with some of the SARS-CoV-2 variants now circulating. This result can also be obtained within an hour, potentially making it much easier to detect different variants of the virus, especially in regions that do not have access to genetic sequencing facilities.
“We have shown that our platform can be programmed to detect new variants that emerge, and that we can reuse it quite quickly,” said James Collins, the Termeer Professor of Medical Engineering and Science in MIT’s Institute for Medical Engineering and Science (IMES ) and Department of Biological Engineering. “In this study, we focused on the British, South African and Brazilian variants, but you could easily adapt the diagnostic platform to the Delta variant and others that are emerging.”
The new diagnosis, which is based on CRISPR technology, can be assembled for about $15, but those costs could drop significantly if the devices were produced on a large scale, the researchers say.
Collins is the senior author of the new study, which appears today (Aug. 6, 2021) in Science Advances. The paper’s lead authors are Helena de Puig, a postdoctoral fellow at Harvard University’s Wyss Institute for Biologically Inspired Engineering; Rose Lee, an instructor of pediatrics at Boston Children’s Hospital and Beth Israel Deaconess Medical Center and a visiting fellow at the Wyss Institute; Devora Najjar, a graduate student in MIT’s Media Lab; and Xiao Tan, a clinical fellow at the Wyss Institute and an instructor in gastroenterology at Massachusetts General Hospital.
A standalone diagnosis
The new diagnosis is based on SHERLOCK, a CRISPR-based tool that Collins and others first reported in 2017. Components of the system include an RNA guide strand that allows for detection of specific target RNA sequences, and Cas enzymes that splicing sequences and a fluorescent signal. All of these molecular components can be freeze-dried for long-term storage and reactivated when exposed to water.
Last year, Collins’ lab began adapting this technology to detect the SARS-CoV-2 virus, in the hopes that they could design a diagnostic device that could provide rapid results and be tested with little or no expertise. served. They also wanted it to work with saliva samples, making it even easier for users.
To do that, the researchers had to include a crucial preprocessing step that knocks out enzymes called salivary nucleases, which destroy nucleic acids like RNA. Once the sample enters the device, the nucleases are inactivated by heat and two chemical reagents. Next, viral RNA is extracted and concentrated by passing the saliva through a membrane.
“That membrane was key to collecting the nucleic acids and concentrating them so we can get the sensitivity we’re showing with this diagnosis,” Lee says.
This RNA sample is then exposed to freeze-dried CRISPR/Cas components, which are activated by automatically puncturing sealed water packets in the device. The one-pot reaction amplifies the RNA sample and then detects the target RNA sequence, if any.
“Our goal was to make a completely self-contained diagnosis that doesn’t require any other equipment,” Tan says. “Essentially, the patient spits into this device, and then you push down a plunger and you get a response an hour later.”
The researchers designed the device, which they call minimally instrumented SHERLOCK (miSHERLOCK), so that it can have up to four modules, each looking for a different target RNA sequence. The original module contains RNA guide strands that detect any strain of SARS-CoV-2. Other modules are specific to mutations related to some of the variants that emerged in the past year, including B.1.1.7, P.1, and B.1.351.
The Delta variant was not yet widespread when the researchers conducted this study, but since the system is already built, they say it should be easy to design a new module to detect that variant. The system can also be easily programmed to check for new mutations that could make the virus more contagious.
“If you want to do a broader epidemiological survey, you can design tests before a mutation of concern occurs in a population, to check for potentially dangerous mutations in the spike protein,” says Najjar.
Follow variants
The researchers tested their device first with human saliva spiked with synthetic SARS-CoV-2 RNA sequences, and then with about 50 samples from patients who tested positive for the virus. They found that the device was as accurate as the gold standard PCR tests in use today, which require nasal swabs and require more time and significantly more hardware and sample handling to provide results.
The device produces a fluorescent readout that can be seen with the naked eye, and the researchers have also designed a smartphone app that can read the results and send them to public health departments for easier tracking.
The researchers believe their device can be produced at a cost of as little as $2 to $3 per device. If approved by the FDA and produced on a large scale, they think this kind of diagnostic could be useful, either for people who want to be able to test at home, or in health centers in areas without widespread access to PCR testing or genetic sequencing of SARS. -CoV-2 variants.
“The ability to detect and track these variants is essential for effective public health, but unfortunately, variants are currently only diagnosed by nucleic acid sequencing in specialized epidemiological centers that are scarce even in resource-rich countries,” says de Puig.
Reference; August 6, 2021, Science Progress.
DOI: 10.1126 / sciaadv.abh2944
The research was funded by the Wyss Institute; the Paul G. Allen Frontiers group; the Harvard University Center for AIDS Research, which is supported by the National Institutes of Health; a Burroughs-Wellcome American Society of Tropical Medicine and Hygiene postdoctoral fellowship; an American Gastroenterological Association Takeda Pharmaceutical Research Scholar Award; and an MIT-TATA Center scholarship.










