New “Atlas” Charts How Antibodies Attack SARS-CoV-2 Coronavirus Spike Protein Variants
0 View
Share this Video
- Publish Date:
- 30 July, 2021
- Category:
- Covid
- Video License
- Standard License
- Imported From:
- Youtube
Tags
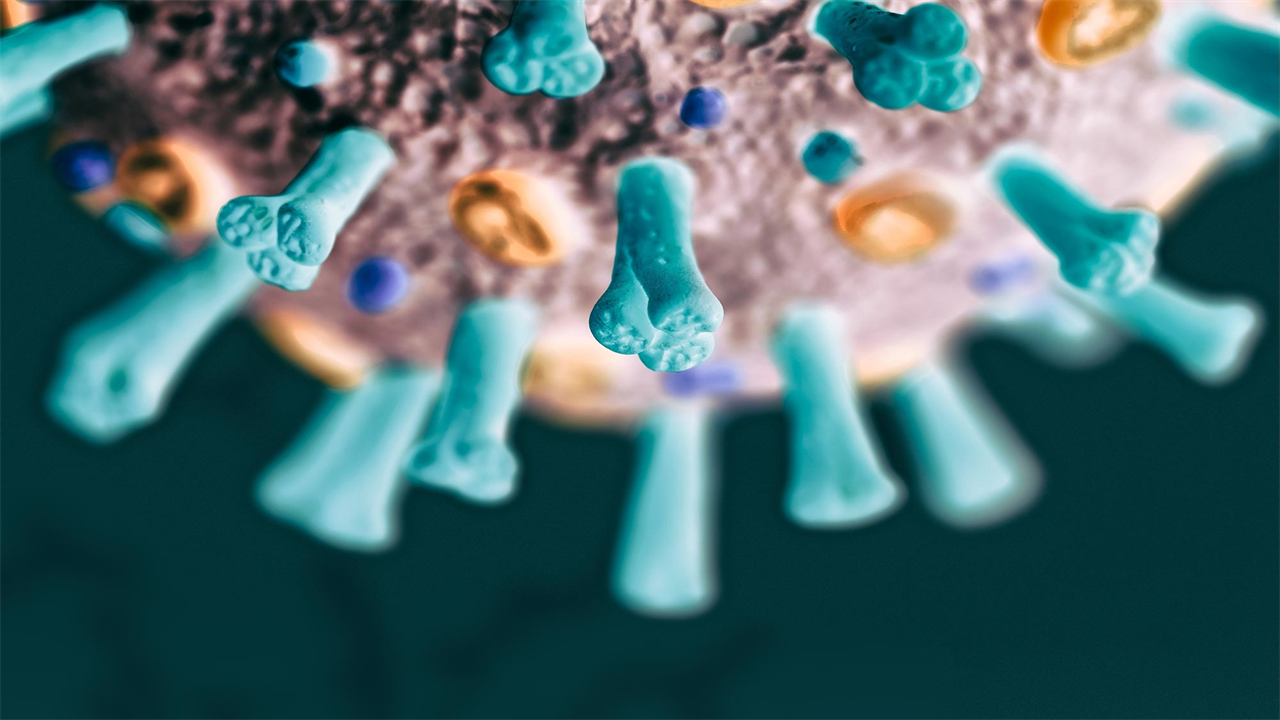
Antibodies capable of neutralizing multiple SARS-CoV-2 strains may inform strategies for overall protective COVID-19 booster vaccines.
As the SARS-CoV-2 virus that causes COVID-19 continues to evolve, immunologists and infectious disease experts are eager to find out if new variants are resistant to the human antibodies that the first versions of the virus recognized. Vaccines against COVID-19, developed from the chemistry and genetic code of this original virus, may offer less protection if the antibodies they help people produce don’t ward off new virus strains. Now, researchers from Brigham and Women’s Hospital and collaborators have created an “atlas” charting how 152 different antibodies attack an important piece of the SARS-CoV-2 machinery, the spike protein, as it has evolved since 2020. Their study, published in Cell, highlights antibodies capable of neutralizing the newer strains while identifying areas of the spike protein that have become more resistant to attack.
“Emerging data shows that vaccines still offer some protection against new SARS-CoV-2 variants, and our study shows how that works from an antibody standpoint,” says corresponding author Duane Wesemann, MD, PhD, of the Department of Allergy and Clinical Immunology and Department of Genetics at Brigham and an associate professor at Harvard Medical School. “These data can help us think about what the best kind of booster vaccine might be by studying how the repertoire of human antibodies recognizes the spike protein.”
The researchers examined the antibody-producing Memory B cells of 19 patients infected with SARS-CoV-2 in March 2020, before new variants emerged. They studied how these antibodies, as well as other antibodies characterized by researchers, bind to spike protein models of the B.1.1.7 (Alpha), B.1351 (Beta), and P.1 (Gamma) variants of SARS. -CoV-2, which were first identified in the United Kingdom, South Africa and Brazil, respectively. An analysis of the Delta variant is currently being worked on.
Overall, the authors confirmed that the hundreds of antibodies they studied largely bind to seven major “footprints” on the spike protein. While many of these antibodies “compete” to bind to the same regions of the early version of the SARS-CoV-2 spike protein, when it comes to newer strains, some of these antibodies lose their potency, while others show up as common. reacting neutralizers.
In particular, antibodies that bind to two of these spike protein regions, named RBD-2 and NTD-1, were the most potent neutralizers of initial forms of the spike protein. The B.1.351 spike variant was found to have the greatest ability to evade existing antibody arsenals and escaped many RBD-2 and NTD-1 binding antibodies. Some antibodies that bind to a different region, called S2-1, can recognize spike proteins from more closely related viruses such as MERS, SARS and cold coronaviruses.
“Making different antibodies that compete for one region of the virus allows the immune system to be more flexible,” Wesemann said. “Otherwise redundant recognition by antibodies targeting the same footprint of one version of the virus gives recognition depth of the same footprint to variants, and some antibodies maintain high neutralization power against all variants. Now that we can identify the antibodies that respond more broadly to all variants.” , we can think about how we can make them stronger in a vaccine.”
Reference: “Memory B Cell Repertoire for Recognition of Evolving SARS-CoV-2 Spike” Pei Tong, Avneesh Gautam, Ian W. Windsor, Meghan Travers, Yuezhou Chen, Nicholas Garcia, Noah B. Whiteman, Lindsay GA McKay, Nadia Storm, Lauren E. Malsick, Anna N. Honko, Felipe JN Lelis, Shaghayegh Habibi, Simon Jenni, Yongfei Cai, Linda J. Rennick, W. Paul Duprex, Kevin R. McCarthy, Christy L. Lavine, Teng Zuo, Junrui Lin, Adam Zuiani, Jared Feldman, Elizabeth A. MacDonald, Blake M. Hauser, Anthony Griffths, Michael S. Seaman, Aaron G. Schmidt, Bing Chen, Donna Neuberg, Goran Bajic, Stephen C. Harrison, Duane R. Wesemann, Accepted, Cell .
DOI: 10.116/j.cell.2021.07.025
This study was supported by NIH grants T32 AI007245, T32 GM007753, AI146779, AI007512, T32 AI007306, AI121394, AI139538 and AI137940, and by MassCPR and Fast Grants for COVID Science.










