Dynamic Model of SARS-CoV-2 Spike Protein Reveals Potential New COVID Vaccine Targets
0 View
Share this Video
- Publish Date:
- 12 April, 2021
- Category:
- Covid
- Video License
- Standard License
- Imported From:
- Youtube
Tags
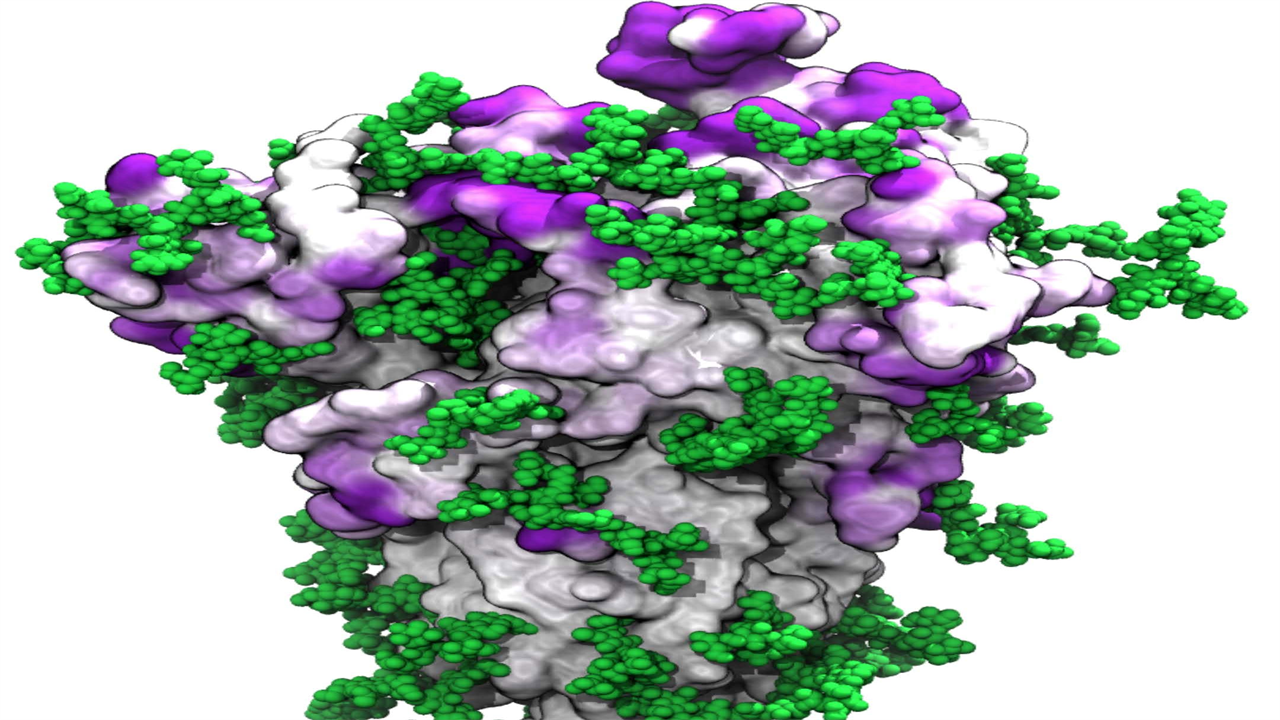
In this visualization of antibody target sites, the SARS-CoV-2 spike protein is attached to the viral membrane with a thin stem. Patches of intense purple color on the surface of the peak indicate potential target sites for antibodies that are not protected by the glycans – chains of sugar molecules – shown in green. These binding sites and their accessibility were identified with molecular dynamics simulations that capture the entire structure of the spike protein and its movements in a realistic environment. Credit: Mateusz Sikora, Sören von Bülow, Florian EC Blanc, Michael Gecht, Roberto Covino, and Gerhard Hummer
New model captures glycan molecules whose movements protect much of the peak from immune defenses.
A new, detailed model of the surface of the SARS-CoV-2 spike protein reveals previously unknown vulnerabilities that could inform vaccine development. Mateusz Sikora of the Max Planck Institute of Biophysics in Frankfurt, Germany, and colleagues present these findings in the open access journal PLOS Computational Biology.
SARS-CoV-2 is the virus responsible for the COVID-19 pandemic. An important feature of SARS-CoV-2 is the spike protein, which extends from the surface and allows it to target and infect human cells. Extensive research has resulted in detailed static models of the spike protein, but these models do not capture the flexibility of the spike protein itself, nor the movements of protective glycans – chains of sugar molecules – that envelop it.
To support vaccine development, Sikora and colleagues sought to identify new potential target sites on the surface of the spike protein. To do this, they developed molecular dynamics simulations that capture the entire structure of the spike protein and its movements in a realistic environment.
These simulations show that glycans on the spike protein act as a dynamic shield that helps the virus evade the human immune system. As with car wipers, the glycans cover almost the entire surface of the nail by flopping back and forth, even though their coverage is minimal at all times.
By combining the dynamic spike protein simulations with bioinformatic analysis, the researchers identified spots on the surface of the spike proteins that are least protected by the glycan shields. Some of the sites detected have been identified in previous research, but some are new. The vulnerability of many of these new sites was confirmed by other research groups in later laboratory experiments.
“We are in a phase of the pandemic, driven by the emergence of new variants of SARS-CoV-2, with mutations mainly concentrated in the spike protein,” says Sikora. “Our approach can support vaccine and therapeutic antibody design, especially when established methods are struggling.”
The method developed for this study could also be applied to identify potential vulnerabilities of other viral proteins.
Reference: “Computational Epitope Map of SARS-CoV-2 Spike Protein” by Mateusz Sikora, Sören von Bülow, Florian EC Blanc, Michael Gecht, Roberto Covino and Gerhard Hummer, April 1, 2021, PLOS Computational Biology.
DOI: 10.1371 / journal.pcbi.1008790










