Achilles’ Heel of SARS-CoV-2 Viral RNA Identified
0 View
Share this Video
- Publish Date:
- 26 August, 2021
- Category:
- Covid
- Video License
- Standard License
- Imported From:
- Youtube
Tags
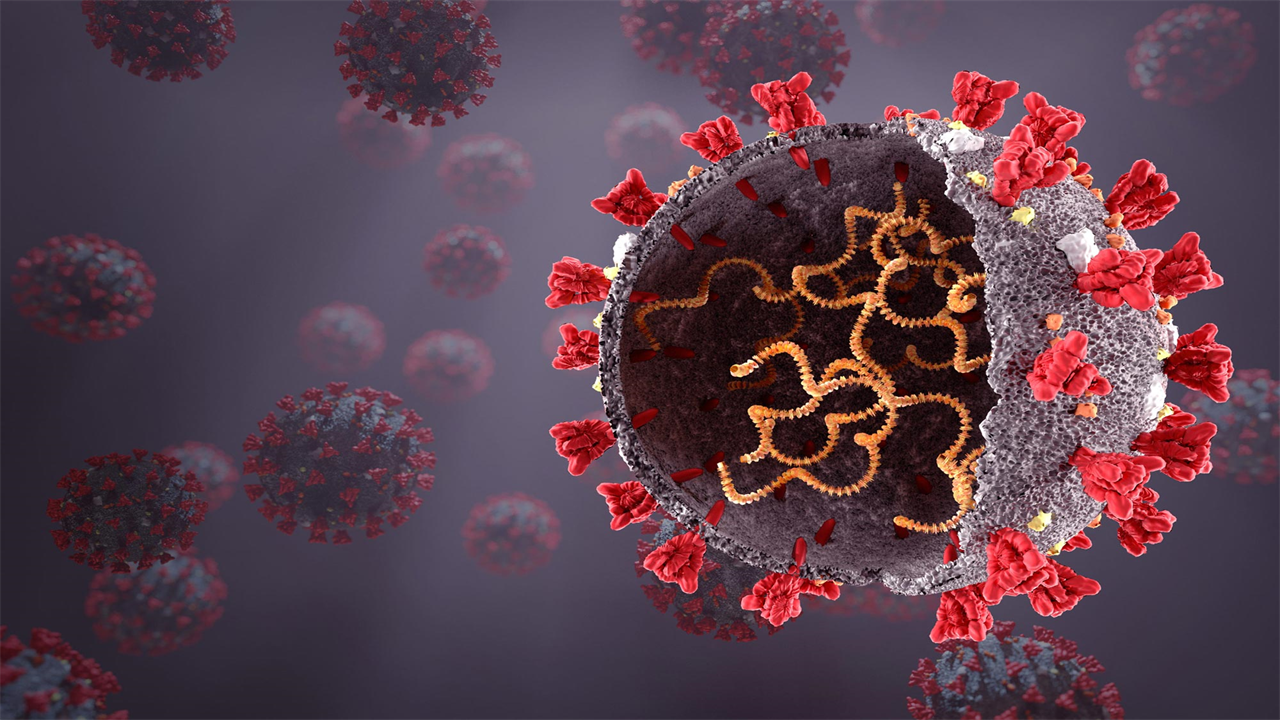
Potential inhibitors of SARS-CoV-2 can not only target viral proteins such as the spike protein (red), but can also act directly on the viral RNA (yellow, in the virus).
Certain regions of the SARS-CoV-2 genome may be a suitable target for future drugs. Researchers from Goethe University, together with their employees in the international COVID-19 NMR consortium, have now discovered this. Using special substance libraries, they have identified several small molecules that bind to certain parts of the SARS-CoV-2 genome that are almost never altered by mutations.
When SARS-CoV-2 infects a cell, it introduces its RNA into it and reprograms it in such a way that the cell produces viral proteins first and then whole viral particles. In the search for active substances against SARS-CoV-2, researchers have so far mainly concentrated on the viral proteins and on blocking them, because this promises to prevent or at least slow down replication. But attacking the viral genome, a long RNA molecule, can also stop or slow viral replication.
The scientists in the COVID-19 NMR consortium, which is coordinated by Professor Harald Schwalbe of the Institute of Organic Chemistry and Chemical Biology at Goethe University, have now taken an important first step in developing such a new class of SARS-CoV -2 drugs. They identified 15 short segments of the SARS-CoV-2 genome that are very similar in several coronaviruses and are known to perform essential regulatory functions. Also in the course of 2020, these segments were very rarely affected by mutations.
The researchers let a substance library of 768 small, chemically simple molecules interact with the 15 RNA segments and analyzed the result using NMR spectroscopy. In NMR spectroscopy, molecules are first labeled with special types of atoms (stable isotopes) and then exposed to a strong magnetic field. The atomic nuclei are excited by means of a short radio frequency pulse and emit a frequency spectrum, with the help of which the RNA and protein structure can be determined and how and where small molecules bind.
This allowed the research team led by Professor Schwalbe to identify 69 small molecules that bound to 13 of the 15 RNA segments. Professor Harald Schwalbe: “Three of the molecules even bind specifically to only one RNA segment. This allowed us to demonstrate that the SARS-CoV-2 RNA is very suitable as a potential target structure for drugs. Given the large number of SARS-CoV-2 mutations, such conservative RNA segments, such as the one we identified, are particularly interesting for developing potential inhibitors. And since the viral RNA makes up up to two-thirds of all RNA in an infected cell, we should be able to significantly disrupt viral replication by using appropriate molecules.”
Against this background, Schwalbe continues, the researchers have already started follow-up tests with readily available substances that are chemically similar to the binding partners from the substance library.
Reference: “Investigating the drugability of conserved RNA regulatory elements in the SARS-CoV-2 genome” by Dr. Sridhar Sreeramulu, Dr Christian Richter, Hannes Berg, Maria A. Wirtz Martin, Betül Ceylan, Tobias Matzel, Jennifer Adam, Nadide Altincekic, Dr. Kamal Azzaoui, Jasleen Kaur Bains, Dr. Marcel JJ Blommers, dr. Jan Ferner, dr. Boris Furtig, prof.dr. Michael Göbel, J. Tassilo Grun, Dr. Martin Hengesbach, Katharina F. Hohmann, Daniel Hymon, Bozana Knezic, Jason N. Martins, Klara R. Mertinkus, Dr. Anna Niesteruk, Stephen A. Peter, Dennis J. Pyper, Dr. Nusrat S. Qureshi, Dr Ute Scheffer, Dr Andreas Schlundt, Dr Robbin Schnieders, Elke Stirnal, Alexey Sudakov, Alix Tröster, Jennifer Vögele, Dr. Anna Wacker, dr. Julia E. Weigand, dr. Julia Wirmer-Bartoschek, prof.dr. Jens Wöhnert and prof.dr. Harald Schwalbe, June 23, 2021, Angewandte Chemie International Edition.
DOI: 10.1002 / anie.202103693










