A COVID Diagnostic in Only 20 Minutes, Using Two CRISPR Enzymes
0 View
Share this Video
- Publish Date:
- 6 August, 2021
- Category:
- Covid
- Video License
- Standard License
- Imported From:
- Youtube
Tags
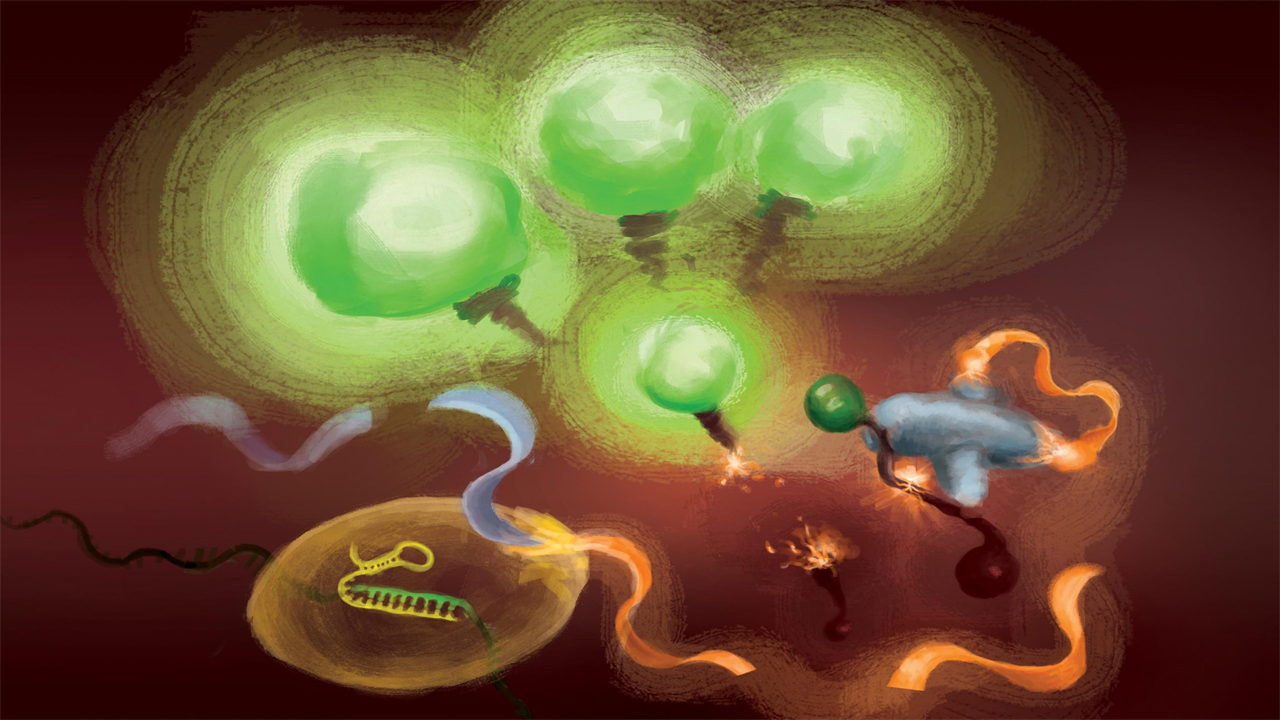
The reactions involved in the FIND-IT test to detect infection with the SARS-CoV-2 virus. When the Cas13 enzyme (left) binds to its target RNA, it cleaves a molecule (orange and gray) to release an activator (orange) that drives the Csm6 nuclease (bottom center) to produce fluorescent markers. cleave and release that light up (green) and signal the presence of viral RNA. Credit: Artwork courtesy of Margaret L. Liu, University of Chicago Pritzker School of Medicine
Frequent, rapid testing for COVID-19 is critical to controlling the spread of outbreaks, especially as new, more transmissible variants emerge.
Although the current diagnostic test for COVID-19, a gold standard, using qRT-PCR – quantitative reverse transcriptase polymerase chain reaction (PCR) – is extremely sensitive, detecting only one copy of RNA per microliter, it requires specialized equipment , a runtime of several hours and a centralized laboratory facility. As a result, testing usually takes at least one to two days.
A research team led by scientists in the labs of Jennifer Doudna, David Savage and Patrick Hsu of the University of California, Berkeley, aims to develop a diagnostic test that is much faster and easier to deploy than qRT-PCR. It has now combined two different types of CRISPR enzymes to create a test that can detect small amounts of viral RNA in less than an hour. Doudna shared the 2020 Nobel Prize in Chemistry for the invention of CRISPR-Cas9 genome editing.
While the new technique is not yet at the stage where it can match the sensitivity of qRT-PCR, which can detect only a few copies of the virus per microliter of liquid, it is already able to pick up levels of viral RNA – about 30 copies per microliter – enough to monitor the population and limit the spread of infection.
“You don’t need the sensitivity of PCR to catch and diagnose COVID-19 in the community, if the test is easy enough and fast enough,” said study co-author David Savage, a professor of molecular and cell biology. “Our hope was to take biochemistry as far as possible to the point where you could imagine a really handy format in an environment where you can be tested every day, for example at the entrance of work.”
The researchers reported their results on Aug. 5, 2021 in the journal Nature Chemical Biology.
Tina Liu and Jennifer Doudna outside the IGI Building on the day Doudna won the 2020 Nobel Prize in Chemistry. Credit: UC Berkeley photo by Brittany Hosea-Small
Several CRISPR-based tests have been approved for emergency use by the Food and Drug Administration, but all require an initial step in which the viral RNA is amplified so that the detection signal — releasing a fluorescent molecule that glows under blue light — is bright enough to to see. While this initial amplification increases the sensitivity of the assay to a similar level to qRT-PCR, it also introduces steps that make it more difficult to perform the assay outside of a lab.
The UC Berkeley-led team tried to achieve useful sensitivity and speed without sacrificing the simplicity of the test.
“For point-of-care applications, you want to be able to react quickly so that people know quickly whether they are infected or not, for example before you get on a flight or visit relatives,” said team leader Tina Liu, a research scientist in the lab of Doudna at the Innovative Genomics Institute (IGI), a CRISPR-focused center involving scientists from UC Berkeley and UC San Francisco.
Apart from an extra step, another disadvantage of initial amplification is that because it makes billions of copies of viral RNA, there is a greater chance of cross-contamination between patient samples. The new technique developed by the team reverses this and amplifies the fluorescent signal instead, eliminating a major source of cross-contamination.
The amplification-free technique, which they call Fast Integrated Nuclease Detection In Tandem (FIND-IT), would enable fast and inexpensive diagnostic tests for many other infectious diseases.
“While we started this project with the express purpose of impacting COVID-19, I think this particular technique could be applicable to more than just this pandemic, because CRISPR is ultimately programmable,” said Liu. “So you could load the CRISPR enzyme with a sequence that targets the flu virus or the HIV virus or any type of RNA virus, and the system has the potential to work the same way. This article really establishes that this biochemistry is a simpler way to detect RNA and has the ability to detect that RNA in a sensitive and rapid time frame that could be amenable to future applications in point-of-care diagnostics.”
The researchers are currently building such a diagnosis using FIND-IT, which includes steps to collect and process samples and to run the test on a compact microfluidic device.
Using tandem Cas proteins
To remove target amplification from the equation, the team used a CRISPR enzyme — Cas13 — to first detect the viral RNA, and another type of Cas protein, called Csm6, to amplify the fluorescent signal.
Cas13 are general purpose scissors for cutting RNA; once it binds to its target sequence, specified by a guide RNA, it is ready to cut a wide variety of other RNA molecules. This target-triggered cutting activity can be used to couple the detection of a specific RNA sequence to the release of a fluorescent reporter molecule. However, on its own, Cas13 can take hours to generate a detectable signal when very low amounts of target RNA are present.
Liu’s insight was to use Csm6 to enhance the effect of Cas13. Csm6 is a CRISPR enzyme that detects the presence of small RNA rings and is activated to cut a wide variety of RNA molecules in cells.
To boost Cas13 detection, she and her colleagues designed a specially designed activator molecule that cuts off when Cas13 detects viral RNA. A fragment of this molecule can bind to Csm6 and prompt it to cut and release a bright fluorescent molecule from a piece of RNA. Normally, the activator molecule is rapidly degraded by Csm6, limiting the amount of fluorescent signal it can generate. Liu and her colleagues devised a way to chemically modify the activator so that it is protected from degradation and can supercharge Csm6 to repeatedly cut and release fluorescent molecules linked to RNA. This results in a sensitivity that is 100 times better than the original activator.
“When Cas13 is activated, it cleaves this small activator and removes a segment that protects it,” Liu said. “Now that it’s released, it can activate many different molecules of that second enzyme, Csm6. And so one target recognized by Cas13 not only triggers activation of its own RNA-cutting ability; it leads to the production of many more active enzymes, each of which can cleave even more fluorescent reporters.”
The team of researchers also incorporated an optimized combination of guide RNAs that allows for more sensitive recognition of the viral RNA by Cas13. When this was combined with Csm6 and its activator, the team was able to detect up to 31 copies per microliter of SARS-CoV-2 RNA in just 20 minutes.
The researchers also added extracted RNA from patient samples to the FIND-IT assay in a microfluidic cartridge, to see if this assay could be adapted to run on a portable device. Using a small device with a camera, they were able to detect SARS-CoV-2 RNA extracted from patient samples with a susceptibility that would capture COVID-19 infections at their peak.
“This tandem nuclease approach — Cas13 plus Csm6 — combines everything in a single reaction at a single temperature, 37 degrees Celsius, so it doesn’t require high temperature heating or multiple steps, as is required for other diagnostic techniques,” Liu said. “I think this opens up opportunities for faster, simpler tests that can achieve comparable sensitivity to other current techniques and potentially achieve even higher sensitivities in the future.”
The development of this amplification-free method of RNA detection was the result of a reorientation of research within IGI as the pandemic began towards problems with the diagnosis and treatment of COVID-19. Ultimately, five UC Berkeley labs and two UCSF labs became involved in this research project, one of many within the IGI.
“When we started this, we had hopes of creating something similar to PCR, but not requiring amplification — that would be the dream,” said Savage, the project’s principal investigator. “And from a sensitivity perspective, we had a gap of about ten thousand times to jump. We made it about a thousandfold; we lowered it about three orders of magnitude. So, we’re almost there. Last April, when we really started mapping it out, that seemed almost impossible.”
Reference: “Accelerated RNA Detection Using Tandem CRISPR Nucleases” by Tina Y. Liu, Gavin J. Knott, Dylan CJ Smock, John J. Desmarais, Sungmin Son, Abdul Bhuiya, Shrutee Jakhanwal, Noam Prywes, Shreeya Agrawal, María Díaz de León Derby, Neil A. Switz, Maxim Armstrong, Andrew R. Harris, Emeric J. Charles, Brittney W. Thornton, Parinaz Fozouni, Jeffrey Shu, Stephanie I. Stephens, G. Renuka Kumar, Chunyu Zhao, Amanda Mok , Anthony T. Iavarone, Arturo M. Escajeda, Roger McIntosh, Shineui Kim, Eli J. Dugan, IGI Testing Consortium, Katherine S. Pollard, Ming X. Tan, Melanie Ott, Daniel A. Fletcher, Liana F. Lareau, Patrick D. Hsu, David F. Savage, and Jennifer A. Doudna, Aug 5, 2021, Nature Chemical Biology.
DOI: 10.1038/s41589-021-00842-2
The work was supported by the Defense Advanced Research Projects Agency (N66001-20-2-4033). Co-authors of the paper include members of the labs of Jennifer Doudna, David Savage, Patrick Hsu, Liana Lareau, and Daniel Fletcher of UC Berkeley; Gavin Knott at Monash University in Australia; Melanie Ott and Katherine Pollard at Gladstone Institutes and UCSF; and Ming Tan at Wainamics, a research and development company in Pleasanton, California, that manufactures microfluidic devices. Doudna, the founder of IGI and currently Chairman and Chair of the IGI Board of Directors, is the Li Ka Shing Chancellor’s Chair at UC Berkeley and a professor of chemistry, molecular and cell biology. Hsu, Lareau and Fletcher are faculties of the Department of Bioengineering.










